About SimpleSynteny
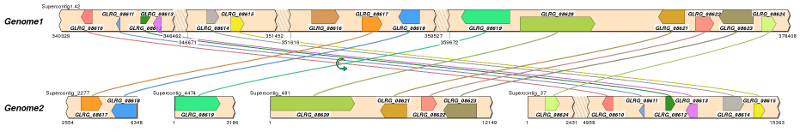
Summary: SimpleSynteny is a graphical program to help biologists easily generate publication-quality visuals for comparative genome analysis or to assist with teaching. The program is designed to help perform a targeted analysis with selected contigs from 1-10 genomes using between 1-60 gene targets. Example uses could be to compare secondary metabolites or look for rearrangements in mating genes across taxa of interest. This implementation of SimpleSynteny is currently not designed to perform annotation for entire (large) genomes and the user should already have a general idea about their genome and gene targets of interest. We recommend users first read the SimpleSynteny paper, available here, from Nucleic Acids Res. (Open Access).
Basic Usage: There are three steps to use the regular program and the user only requires two or more FASTA files to get started.
- Step 1: Upload one or more genome FASTA files (with 1-10 DNA contig/supercontig sequences) and one or more gene FASTA files (with 1-60 DNA or protein sequences) for comparison. If you don't know which contigs in a genome may contain your genes, you can use our 'Contig Finder' program to help identify them and prepare a FASTA file for use with the main program. For each genome in your analysis, enter a desired name and associate the file containing the genes you want to map. You can also adjust additional BLAST settings as needed or, if you are looking at circular or mitochondrial genomes, you may want to enable 'Circular Genome Mode.'
- Step 2: Next, you will manually review the results for each of your genomes. You can reorient ('flip') contigs or remove any unwanted gene fragments that BLAST may have mapped. The program will notify you if any of your genes did not map.
- Step 3: For the last step, you can generate your final image by adjusting any prefered settings (image size, format, resolution, etc.) and download your final results as a zip file.
Output Files: After you download and unzip your final results, you should find your final image in addition to folders containing 'CMAP' (contig mapping) files used to generate your image, your BLAST results, and lists of any genes that were skipped by the pipeline due to no significant hits being found.
Advanced Mode: Want to include additional knowledge to your analysis from other sources? 'Advanced Mode' allows you to manually annotate a figure by submitting a CMAP file to further customize your figure. You can either cut and paste lines from the CMAP files provided by regular SimpleSynteny output as a starting point, or write your own lines from scratch. For example, this mode can be used to manually insert introns/exons, additonal tandem repeats, etc. as separate gene entries.
How it Works: The pipeline utilizes NCBI BLAST, BioRuby and RMagick to map genes onto genomes and generate a final image. The server implementation makes additional use of JQuery and PHP and requires a modern HTML5 compliant browser in order to run.
Questions / Help: Having trouble? Check out the Demo Mode, the introductory flyer, or the Frequently Asked Questions (FAQ) section. Further questions, comments, suggestions or problems using the site can be directed to Dan Veltri . Please include "SimpleSynteny" in the subject line and mention any error message if you encountered one while using the site.
How to Cite SimpleSynteny: Veltri, D., Malapi-Wight, M. and Crouch J.A. SimpleSynteny: a web-based tool for visualization of microsynteny across multiple species. Nucleic Acids Res. 44(W1):W41-W45, 2016, doi:10.1093/nar/gkw330.
Version: The current version of the SimpleSynteny server is v1.6 as of February 22nd, 2023.
Download: SimpleSynteny scripts are available to run locally for larger jobs. Click here to download (v1.4.0 .ZIP 469kB) updated on August 16th, 2023. Previous versions are available here.
Acknowledgments: Support for this project is made possible through funding provided by the 2013-2015 USDA-APHIS Farm Bill 10201 and 10007 Programs; The U.S. Department of Agriculture Agricultural Research Service (USDA-ARS); the USDA-ARS Floriculture and Nursery Research Initiative with the sponsorship of AmericanHort and the Society of American Florists; Rutgers University Department of Plant Biology and Pathology; and the ARS Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the USDA. ORISE is managed by ORAU under DOE contract number DE-AC05-06OR23100.
